| ID |
Date |
Author |
Category |
Subject |
Year |
|
471
|
Wed May 26 15:30:03 2021 |
Ragan, Thanassis | General | Changing the beam energy | 2021 |
The ESR team is changing the beam energy to 6 MeV/u. |
|
316
|
Thu May 6 11:03:46 2021 |
Jan Glorius | DAQ | Cabling Documentation | 2021 |
|
| Attachment 1: e127b_cabling_docu_06_05_2021_.pdf
|
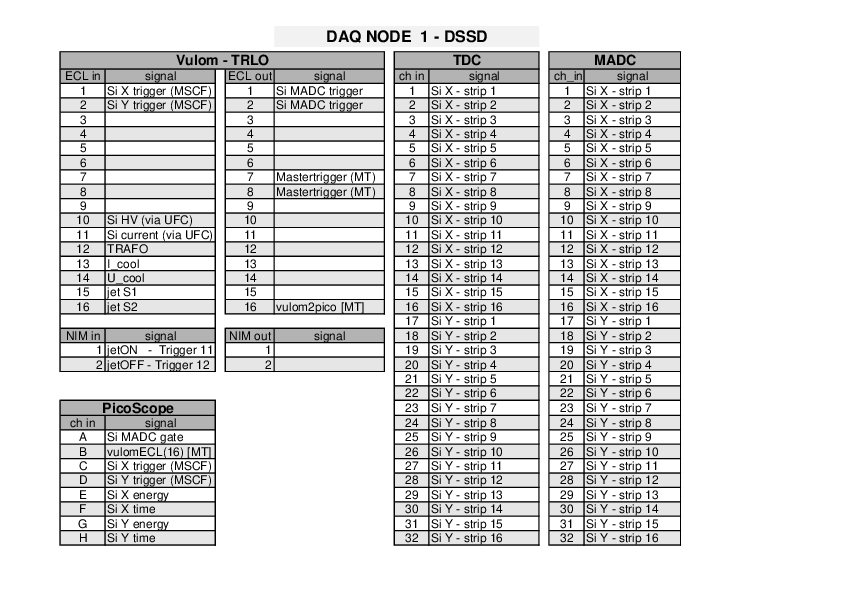

|
| Attachment 2: e127b_2021_cabling_documentation.xlsx
|
|
346
|
Sun May 16 13:31:49 2021 |
Laszlo | DAQ | Cabling Documentation | 2021 |
Si HV and current was switched in the last documentation. |
| Attachment 1: e127b_2021_cabling_documentation.xlsx
|
|
Draft
|
Fri Jun 28 14:06:28 2019 |
Jan | General | CAD of (p,g) setup | |
|
|
350
|
Tue May 18 21:08:16 2021 |
ESR team | General | Beam stacking | 2021 |
Beam stacking. 3 pulses, stochastically cooled and stacked on an inner orbit |
| Attachment 1: 5B6943AF-DF56-4B0F-A77E-1E3C65B3B0C0.jpeg
|
|
| Attachment 2: B590F232-450A-4ACF-BBA4-DB31B7B35FDA.jpeg
|
|
|
572
|
Tue Jun 8 15:06:17 2021 |
Shahab | Analysis | Beam lifetime | 2021 |
measured beam lifetime after the second deceleration using parallel plate Schottky detector and the RSA5103 spectrum analyser. Analysis code can be found here:
https://github.com/xaratustrah/e127_scripts/
The plots here were produced using the script:
https://nbviewer.ipython.org/github/xaratustrah/e127_scripts/blob/main/lifetime_calculator.ipynb
UPDATE 2022-06-30:
Life times have been slightly corrected. Please see attachment.
Filename Start Frame Correct Lifetime [s] Mean [s]
pgamma_lifetime-2021.05.25.21.56.37.593_on.tiq 29500 1.43
pgamma_lifetime-2021.05.25.22.02.16.618_on.tiq 29500 1.55 1.49
pgamma_lifetime-2021.05.25.22.07.22.369_off.tiq 53500 1.75
pgamma_lifetime-2021.05.25.22.13.24.787_off.tiq 34900 1.89 1.82 |
| Attachment 1: pgamma_lifetime.001.png
|
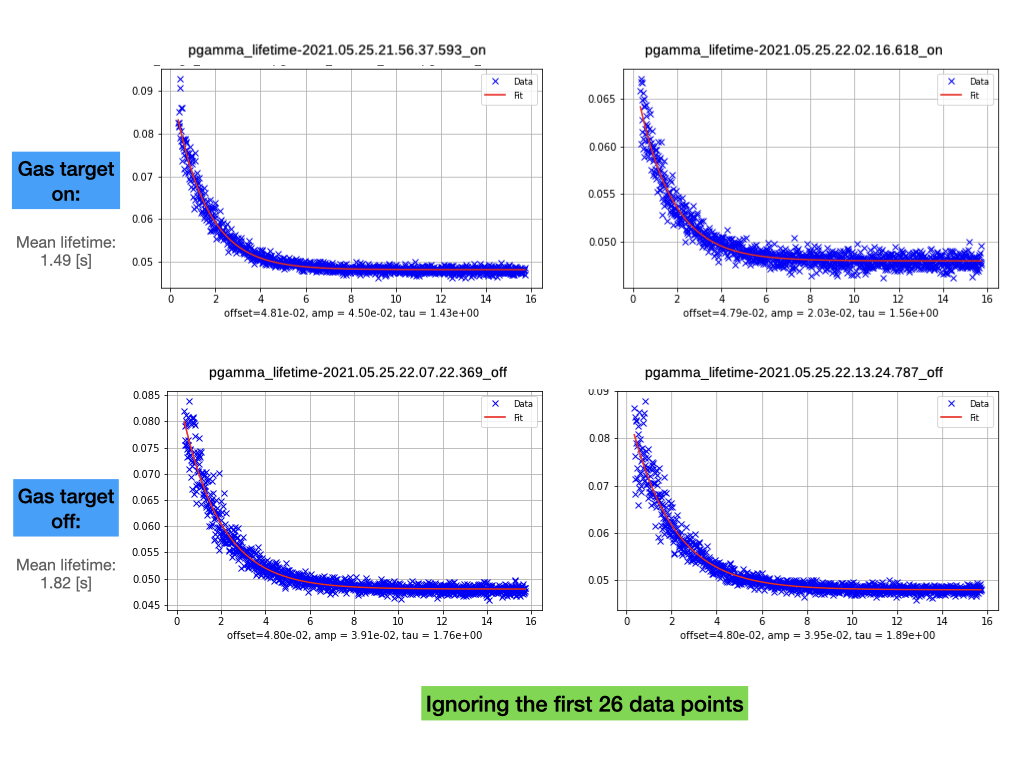
|
| Attachment 2: pgamma_lifetime.002.png
|
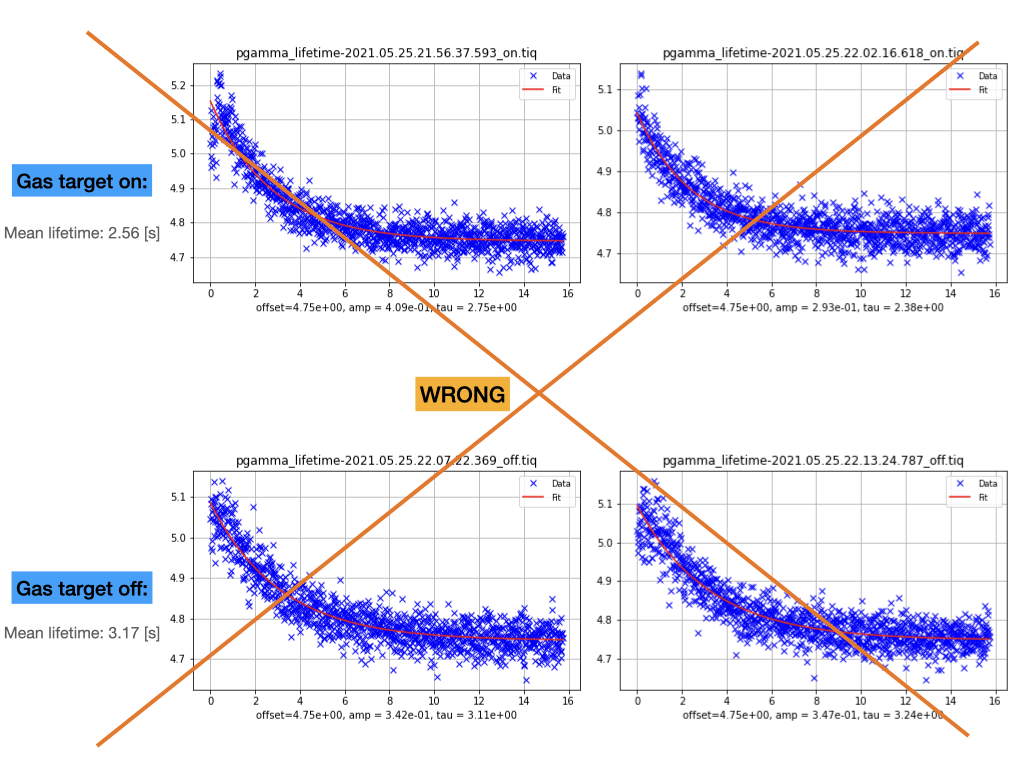
|
|
353
|
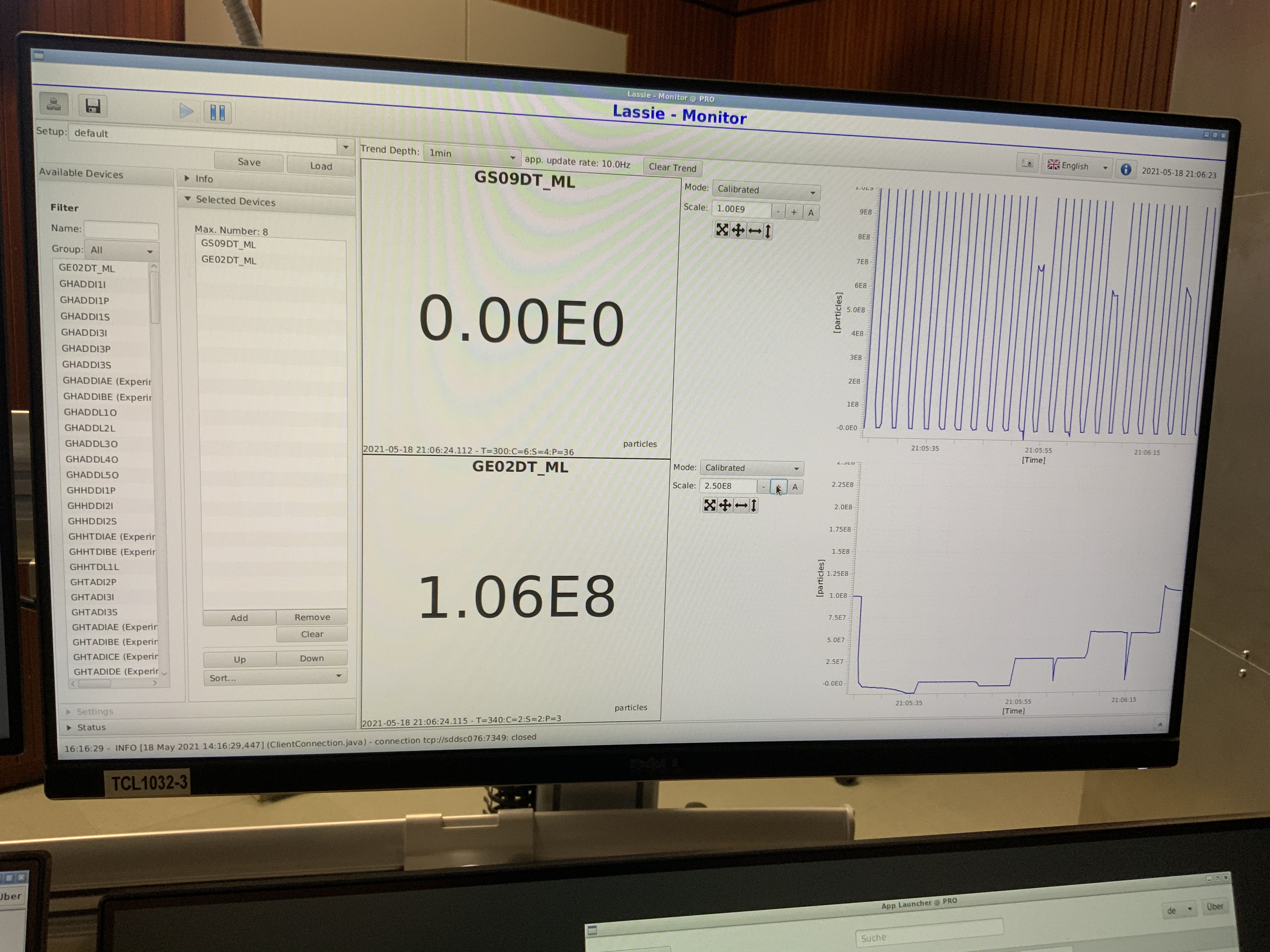
Wed May 19 19:47:14 2021 |
Yuri | General | Beam Halflife at 30 AMeV | 2021 |
|
| Attachment 1: 2021-05-19_17-35-00_tcl1032_lassie-monitor.png
|
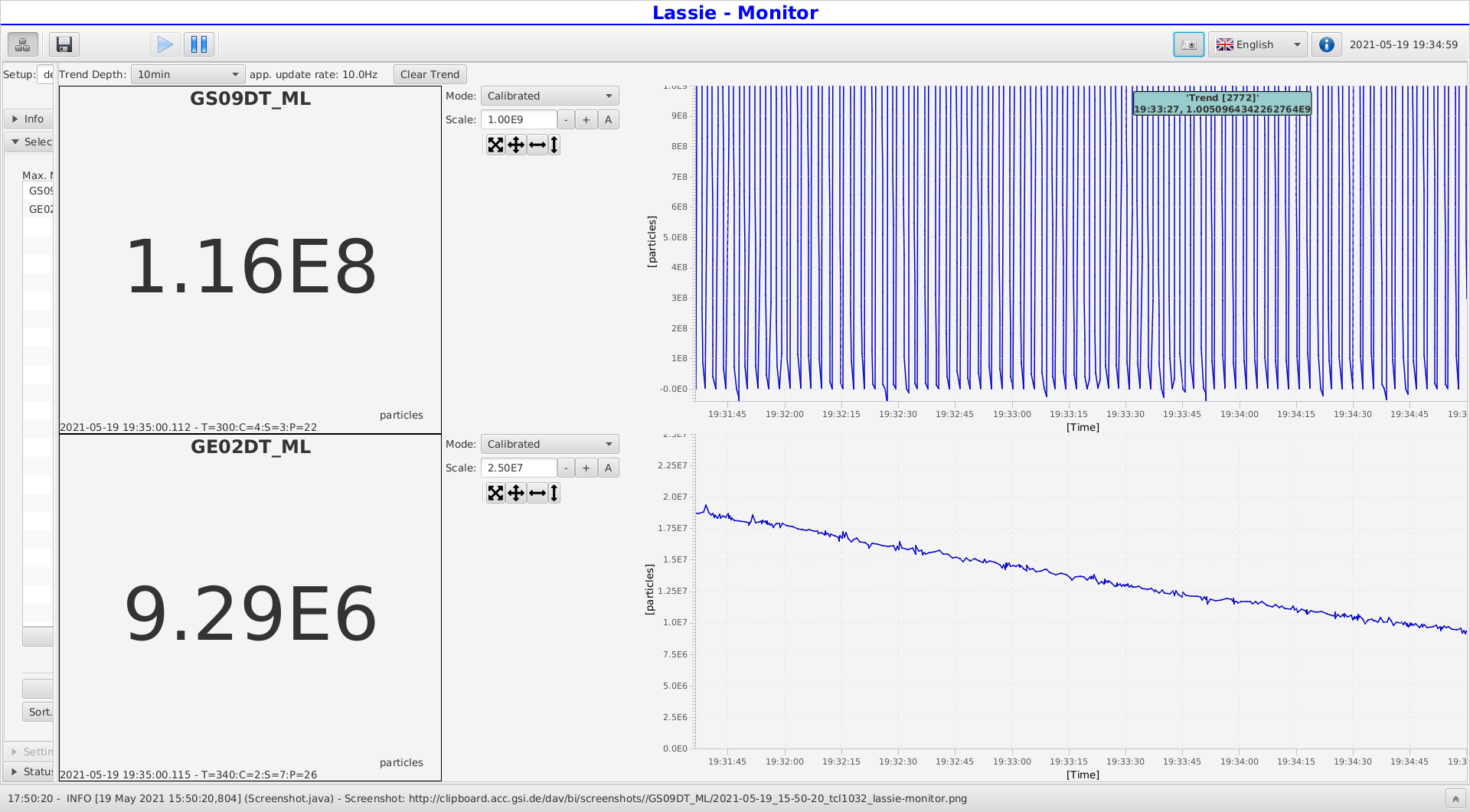
|
|
449
|
Tue May 25 17:12:59 2021 |
Robert/Shahab | Accelerator | Beam | 2021 |
Beam is back; It is now being optimized |
|
250
|
Mon Mar 23 14:23:01 2020 |
Pierre-Michel | DAQ | Backup recoil files | |
Recoil detector files backup to lxg1260:/data.local2/hillenbrand/Laszlo |
|
3
|
Wed Feb 6 10:04:23 2019 |
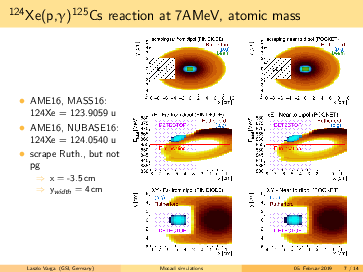
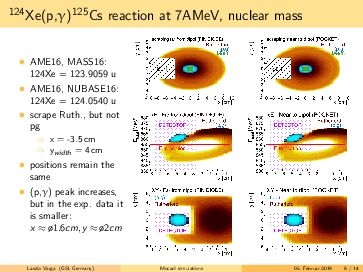
Laszlo Varga | Simulations | Background subtraction for 124Xe, 109In, 73As made by MOCADI | |
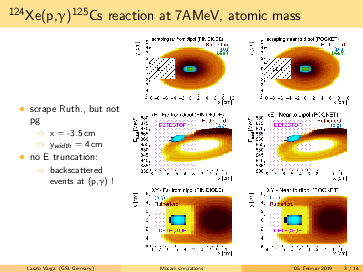
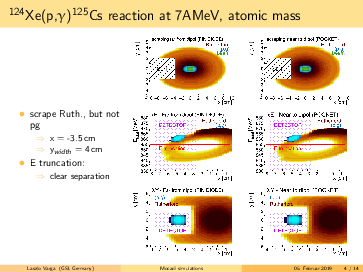
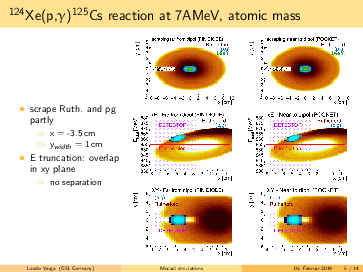
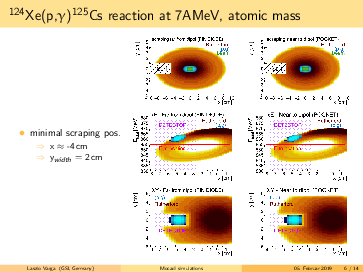
General remarks to the MOCADI simulations:
- scraper size vertically > (p,g) spot at the scraping position --> (p,g) can be separated by truncating the
backscattered Rutherford events in the energy.
- replacing the atomic masses to nuclear masses does not change significantly the absolute positions of the
(p,g) and the Rutherford. However, the (p,g) spot in the simulation seems bigger using the nuclear masses, than
in the 124Xe experiment with a factor ~1.6.
- going for lighter elements, the separation between (p,g) and Rutherford getting better, than slit can be
placed even more far from the beam in x (radial) direction. For 124Xe the minimal distance from the beam axis is
<4cm, for 73As it is <7.5cm.
- in the simulations the scraping is mostly until x=-inf. However, if the scraping is incomplete in x, we can end
up with underlying background events below the p,g peak (see the last slides)! The Rutherford cone gets bigger by
going down with A,Z. For 91Nb the minimal width of the scraper: x>6cm, for 73As x>7cm. Therefore, I suggest to
have a scraper width x=9cm. For the y, y=6cm should be safe.
- for lighter elements, the (p,g) spot size increases: in the simulation for 73As it reaches the detector size.
However, the (p,g) spot size might be overestimated, please read the 2. point.
|
| Attachment 1: template.pdf
|
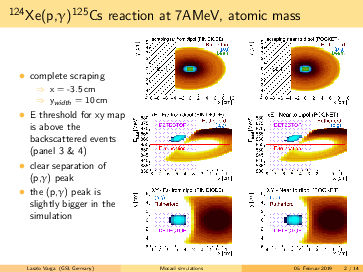
|
|
88
|
Fri Mar 20 22:26:42 2020 |
Jan | Detectors | BaF2 deactivated | |
BaF2 detectors are deactivated nor for the following runs. |
|
32
|
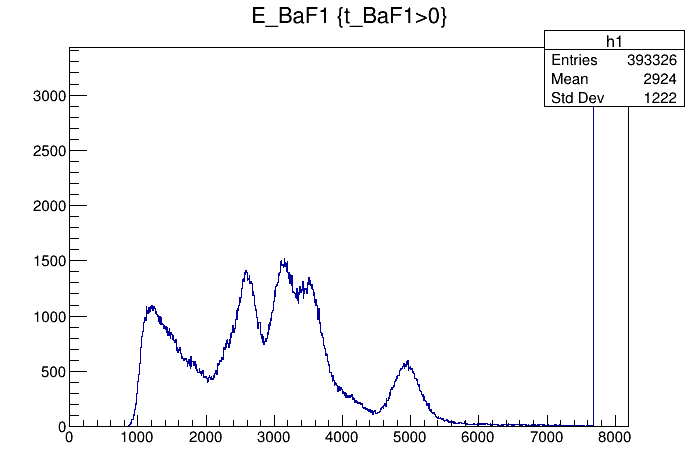
Tue Feb 11 13:20:01 2020 |
Jan | Detectors | BaF2 - HV settings | |
with these HV values the 6 channels are roughly gain matched, such that the 7.83 MeV Po-decay is around channel 5000. |
| Attachment 1: BaF2_HV_settings.png
|
|
| Attachment 2: baf_bg_ch1.png
|
|
|
85
|
Fri Mar 20 16:54:53 2020 |
Jan | DAQ | BaF OFF downscaled red=32 | |
I introduced a downscaling by a factor of 2^5=32 for the BaF2 during target OFF.
The reason is, that we get a lot reasonable x-ray counts (K-alpha, K-REC) outside target ON phase.
So i want to minimize our deadtime during target OFF, to be able to use the data efficiently if needed.
BaF2 is not really needed in target OFF. |
|
288
|
Wed Mar 31 13:52:04 2021 |
Jan | Detectors | BaF Na22 Testing | 2021 |
On 30. and 31.03.2021 we did some testing with the 6 BaF detectors.
Trigger & threshold:
With a threshold at ~250 keV, each detector has an trigger rate of about 0.5kHz due to internal activity.
Energy spectra
All detectors showed the Na22 lines (511 + 1275 keV) and also the lines from internal activity.
The energy resolution was on the order of 8.5 - 11 keV, depending on the detector.
Time spectra
Raw TDC spectra showed self-stop peak.
TDC diff spectra (between detectors) show one main (sharp) correlation peak about 8ns = 30ch*0.293ns/ch off the zero.
At very low intensity a time structure around this peak is visible in a region of 120ns around the main peak. This is not understood, the energy spectra look similar for all these time regions (above/on/below the main peak).
Additionally, the coincidence E-specta between detector 1 and 2 (gating on the main time peak), nicely showed only the 511 keV line from Na22 and some gras.
Overnight test run:
Det. 1, 2, 3, 4 with the Na22 source
lxg1275:/data.local3/e127/lmd_2021/test/
e127b_run0021.lmd
e127b_run0022.lmd
e127b_run0023.lmd
e127b_run0024.lmd
e127b_run0025.lmd
e127b_run0026.lmd
e127b_run0027.lmd
e127b_run0028.lmd
e127b_run0029.lmd
e127b_run0030.lmd
e127b_run0031.lmd
e127b_run0032.lmd
e127b_run0033.lmd
e127b_run0034.lmd
Gain matched HV settings: https://elog.gsi.de/esr/E127/289
Issues to solve:
- 2 missing HV channels
- Go4 Analysis not stable
- trigger not on Baf but on Si |
|
289
|
Wed Mar 31 13:57:27 2021 |
Jan | Detectors | BaF HV settings | 2021 |
The setting below is a start value for final gain matching with a full ADC range of 12MeV:
|
Det.Nr. | ADC/TDC ch | HV | MSCF Gain
| |
1 | 20 | 2400V | 6
| |
2 | 21 | 1780V | 6
| |
3 | 22 | 2350V | 6
| |
4 | 23 | 2130V | 6
| |
5 | 24 | 2050V | 6
| |
6 | 25 | 1780V | 6
|
|
|
290
|
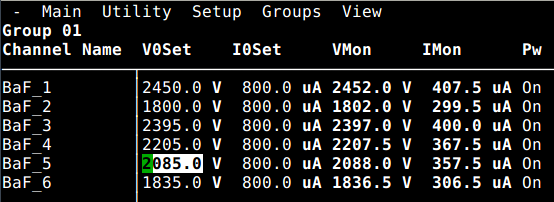
Wed Apr 14 09:42:50 2021 |
Jan | Detectors | BaF HV gain matching | 2021 |
The preliminary gain matching settings for a full ADC range of ~12MeV:
|
Det.Nr. | ADC/TDC ch | HV | I | comment
| |
1 | 20 | 2400V | 390uA | CAEN HV ch0
| |
2 | 21 | 1763V | 287uA | CAEN HV ch1
| |
3 | 22 | 2340V | 380uA | CAEN HV ch2
| |
4 | 23 | 2175V | 354uA | CAEN HV ch3
| |
5 | 24 | 2017.5V | ? | emetron HV
| |
6 | 25 | 1780V | ? | not yet biased
|
Attached is a 22Na-spectrum taken with detectors 1 - 5.
The used MSCF settings are also attached.
Detector 6 does not yet have a HV channel available. |
| Attachment 1: BaF_Na22_gain_match_v1.png
|
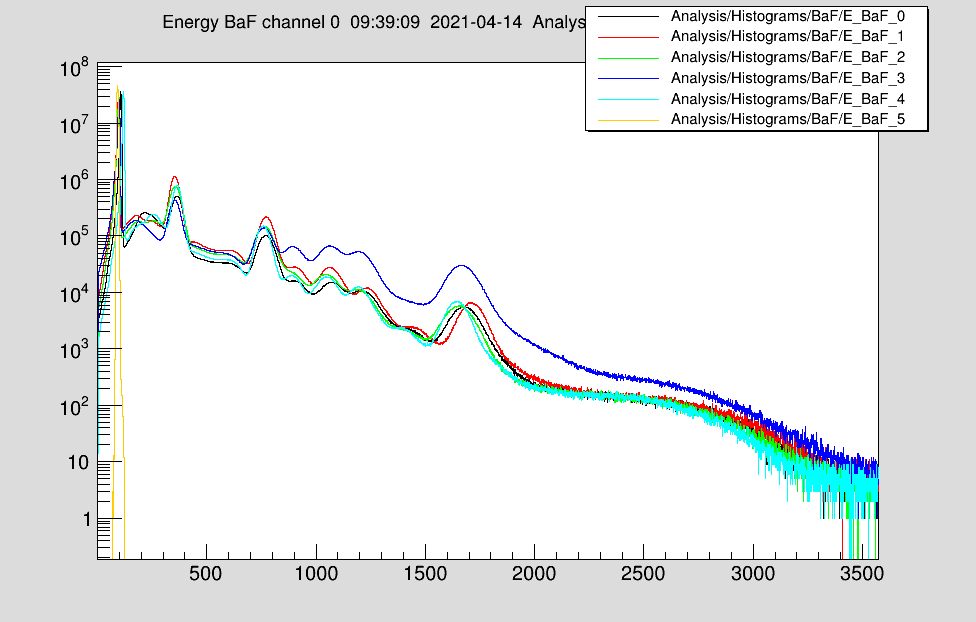
|
| Attachment 2: baf_gain_match_v1.dat
|
atpnuc006:mrcc:mscf1:getGainCommon 15
atpnuc006:mrcc:mscf1:getShapingTimeCommon 3
atpnuc006:mrcc:mscf1:getThresholdCommon 6
atpnuc006:mrcc:mscf1:getPzCommon 6
atpnuc006:mrcc:mscf1:getGain1 6
atpnuc006:mrcc:mscf1:getGain2 6
atpnuc006:mrcc:mscf1:getGain3 6
atpnuc006:mrcc:mscf1:getGain4 6
atpnuc006:mrcc:mscf1:getShapingTime1 3
atpnuc006:mrcc:mscf1:getShapingTime2 3
atpnuc006:mrcc:mscf1:getShapingTime3 3
atpnuc006:mrcc:mscf1:getShapingTime4 3
atpnuc006:mrcc:mscf1:getThreshold1 6
atpnuc006:mrcc:mscf1:getThreshold2 6
atpnuc006:mrcc:mscf1:getThreshold3 6
atpnuc006:mrcc:mscf1:getThreshold4 6
atpnuc006:mrcc:mscf1:getThreshold5 6
atpnuc006:mrcc:mscf1:getThreshold6 6
atpnuc006:mrcc:mscf1:getThreshold7 6
atpnuc006:mrcc:mscf1:getThreshold8 6
atpnuc006:mrcc:mscf1:getThreshold9 6
atpnuc006:mrcc:mscf1:getThreshold10 6
atpnuc006:mrcc:mscf1:getThreshold11 6
atpnuc006:mrcc:mscf1:getThreshold12 6
atpnuc006:mrcc:mscf1:getThreshold13 6
atpnuc006:mrcc:mscf1:getThreshold14 6
atpnuc006:mrcc:mscf1:getThreshold15 6
atpnuc006:mrcc:mscf1:getThreshold16 6
atpnuc006:mrcc:mscf1:getPz1 6
atpnuc006:mrcc:mscf1:getPz2 3
atpnuc006:mrcc:mscf1:getPz3 3
atpnuc006:mrcc:mscf1:getPz4 1
atpnuc006:mrcc:mscf1:getPz5 5
atpnuc006:mrcc:mscf1:getPz6 5
atpnuc006:mrcc:mscf1:getPz7 6
atpnuc006:mrcc:mscf1:getPz8 6
atpnuc006:mrcc:mscf1:getPz9 6
atpnuc006:mrcc:mscf1:getPz10 6
atpnuc006:mrcc:mscf1:getPz11 6
atpnuc006:mrcc:mscf1:getPz12 6
atpnuc006:mrcc:mscf1:getPz13 6
atpnuc006:mrcc:mscf1:getPz14 6
atpnuc006:mrcc:mscf1:getPz15 6
atpnuc006:mrcc:mscf1:getPz16 6
atpnuc006:mrcc:mscf1:getSingleChMode 1
atpnuc006:mrcc:mscf1:getRcMode 1
atpnuc006:mrcc:mscf1:getAutoPZ 0
atpnuc006:mrcc:mscf1:getMultiplicityHi 8
atpnuc006:mrcc:mscf1:getMultiplicityLo 1
atpnuc006:mrcc:mscf1:getSumTrgThresh 100
atpnuc006:mrcc:mscf1:getBlrOn 0
atpnuc006:mrcc:mscf1:getCoincTime 100
atpnuc006:mrcc:mscf1:getThreshOffset 1
atpnuc006:mrcc:mscf1:getShaperOffset 100
atpnuc006:mrcc:mscf1:getBlrThresh 10
atpnuc006:mrcc:mscf1:getECLDelay 0
|
|
521
|
Fri May 28 14:14:31 2021 |
Laszlo | General | Alvarez failure at 10:30 28:05:2021 | 2021 |
|
|
411
|
Sun May 23 15:36:10 2021 |
Rui-Jiu Chen& Alex | General | All shifts until Monday morning (24th May) have been cancelled. | 2021 |
All shifts until Monday morning (24th May) have been cancelled.
Next update on further situation (in particular concerning the morning shift on Monday) will be given tonight . |
|
188
|
Sun Mar 22 13:33:19 2020 |
Laszlo | General | 2nd storage of MCP data | |
I made a backup for the MCP data on my own GSI laptop. Looking for better solution...
the original "fresh" files stored at litv-exp@lxg1299:/data.local2/hillenbrand/Laszlo/ |
|
271
|
Mon Apr 6 04:39:19 2020 |
Laszlo | Calibration | 2. Xray energy calibration | |
30angle: E[keV] = 0.015898 *ch-1.91
90angle: E[keV] = 0.0211075*ch-2.39
145angle: E[keV] = 0.01663 *ch-1.758 |
| Attachment 1: 30angle_Xray_second_calibration.png
|
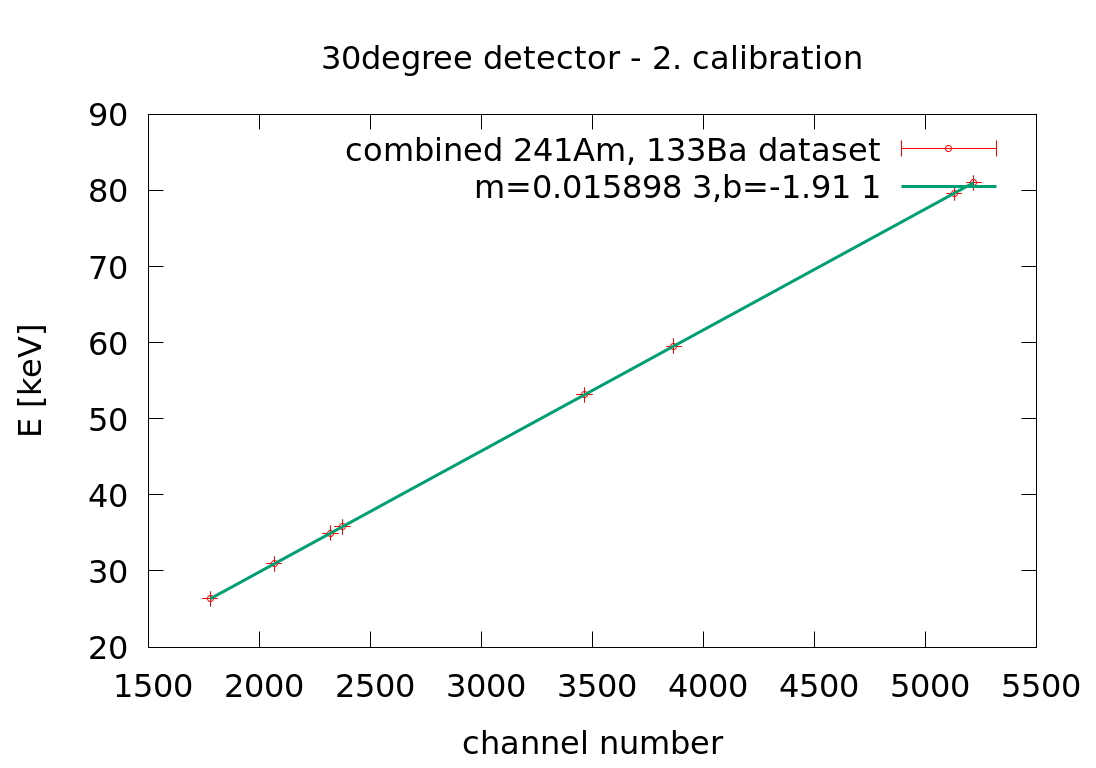
|
| Attachment 2: 90angle_Xray_second_calibration.png
|
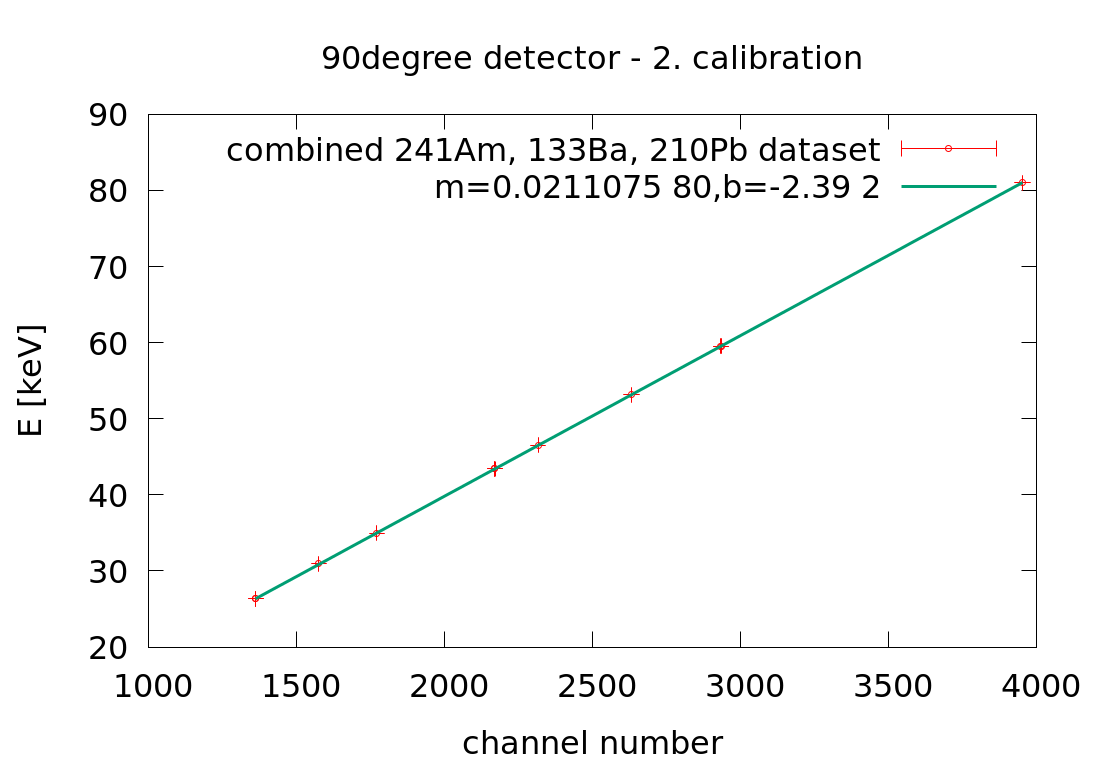
|
| Attachment 3: 145angle_Xray_second_calibration.png
|
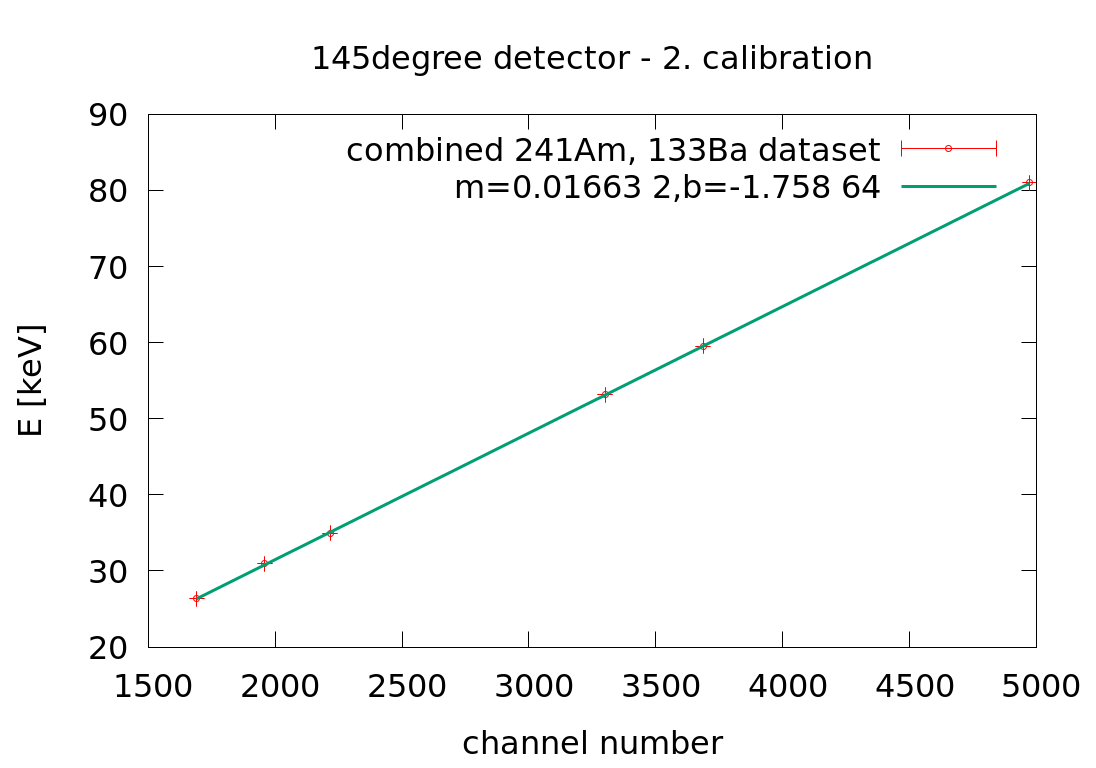
|
| Attachment 4: Am_Ba.cal
|
#gaus fit, gaus fit error, literature (nndc), literature error
#literature used:
#https://www.nndc.bnl.gov/nudat2/decaysearchdirect.jsp?nuc=241AM&unc=nds
#https://www.nndc.bnl.gov/nudat2/decaysearchdirect.jsp?nuc=133BA&unc=nds
#https://www.nndc.bnl.gov/nudat2/decaysearchdirect.jsp?nuc=210PB&unc=nds
#where there is no error given, I put +/-.5 to the last digit
1.77780e+03 8.30485e-02 26.3446 0.0002
3.86555e+03 2.20759e-02 59.5409 0.0001
2.06701e+03 1.25971e-01 30.973 0.0005
2.32011e+03 8.82876e-02 34.987 0.0005
2.37424e+03 2.01021e-01 35.818 0.0005
3.46400e+03 2.53619e-01 53.1622 0.0006
5.12992e+03 2.64070e-01 79.6142 0.0012
5.21686e+03 6.55175e-02 80.9979 0.0011
|
| Attachment 5: Am_Ba.cal
|
#gaus fit, gaus fit error, literature (nndc), literature error
#literature used:
#https://www.nndc.bnl.gov/nudat2/decaysearchdirect.jsp?nuc=241AM&unc=nds
#https://www.nndc.bnl.gov/nudat2/decaysearchdirect.jsp?nuc=133BA&unc=nds
#https://www.nndc.bnl.gov/nudat2/decaysearchdirect.jsp?nuc=210PB&unc=nds
#where there is no error given, I put +/-.5 to the last digit
1.36175e+03 9.00555e-02 26.3446 0.0002
2.16961e+03 8.05264e-01 43.420 0.003
2.93441e+03 2.14830e-02 59.5409 0.0001
1.57547e+03 4.20575e-02 30.973 0.0005
1.77077e+03 1.39155e-01 34.987 0.0005
2.63200e+03 2.99411e-01 53.1622 0.0006
3.95276e+03 6.39294e-02 80.9979 0.0011
1.36182e+03 1.02182e-01 26.3446 0.0002
2.17086e+03 9.05336e-01 43.420 0.003
2.93383e+03 2.43152e-02 59.5409 0.0001
2.31714e+03 3.80548e-01 46.539 0.001
|
| Attachment 6: 145_det_2calib_Am_Ba.cal
|
#gaus fit, gaus fit error, literature (nndc), literature error
#literature used:
#https://www.nndc.bnl.gov/nudat2/decaysearchdirect.jsp?nuc=241AM&unc=nds
#https://www.nndc.bnl.gov/nudat2/decaysearchdirect.jsp?nuc=133BA&unc=nds
#https://www.nndc.bnl.gov/nudat2/decaysearchdirect.jsp?nuc=210PB&unc=nds
#where there is no error given, I put +/-.5 to the last digit
1.69027e+03 2.31158e-01 26.3446 0.0002
3.68596e+03 5.13163e-02 59.5409 0.0001
1.95922e+03 6.96080e-02 30.973 0.0005
2.21589e+03 1.68384e-01 34.987 0.0005
3.30072e+03 5.68458e-01 53.1622 0.0006
4.97302e+03 1.24941e-01 80.9979 0.0011
|
| Attachment 7: run115_133Ba_high_30.svg
|

|
| Attachment 8: run111_241Am_90.svg
|

|
| Attachment 9: run115_210Pb_90.svg
|

|