| ID |
Date |
Author |
Category |
Subject |
Year |
|
576
|
Thu Jul 8 14:07:19 2021 |
Jan | Calibration | beam energies | 2021 | Table of ion energies calculated from cooler parameters
|
nominal energy | V_set | V_meas | I_set | E_ion | E_Error
| |
6 MeV/u | 3460.5 V | 3364 V | 50 mA | 6.042 MeV/u | YY keV/u
| |
7 MeV/u | 4008.3 V | 3913 V | 50 mA | 7.049 MeV/u | YY keV/u
|
Here is how the calculation works:
1. Find the voltage and current set values that were active during the measurement cycle.
2. These are only SetValues. For the current this is fine, but for the voltage you have to subtract an offset. This offset comes from the power supply and changes over time. It is measured regularly and the present offset you have to inquire with the ESR/cooler crew. For our beam time E127b in 2021 the offset is:
V_offset = -95 +/- ?? V (only for E127b)
The uncertainty has still to be discussed.
3. The space charge of the electron beam reduces the effective voltage seen by the electrons and thus reduces their energy. You have to calculate the space charge potential Phi_0 on the beam axis using the following formular:
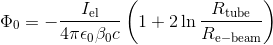
using R_tube = 10cm and R_e-beam = 2,54 cm and all constants, you arrive at the ESR specific equation:
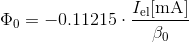
4. Now calculate effective Voltage and electron energy:
V_eff = V_set + V_offset + Phi_0
This time we measured the effective voltage for each energy, so we can actually use this values (V_meas) instead of V_set + V_offset.
E_e = e * V_eff
5. With E_e you can now calculate the ion energy by using the definition of the Lorentzfactor gamma:
gamma - 1 = E_kin/(m_0 * c^2)
In equilibrium, the left side of the equation is equal for both, electrons and ions in the cooler, leading to:
E_ion = E_el * M_ion/M_el
which gives the total energy of the ions.
NOTE:
In principle, this should be an iterative calculation, since the potential depends on beta and is then used to calulate beta or ion energy. But in practice a single iteration is precise enough, if beta was a good estimate. |
|
18
|
Tue Jul 23 10:46:29 2019 |
Laszlo | Analysis | 124Xe pg peak fit using mixed cascade simulation | | This entry is the continuation of the cascade effects on the pg peak shape entry:
https://exp-astro.physik.uni-frankfurt.de:8080/E108/475
The main point is that having more cascade makes the impulse carried by one photon smaller --> recoil cone is
smaller --> pg peak gets more centered
When one wants to make an "all-inklusive" simulation the problem comes, that there are too many populated states
after CN* decay (many are just theoretical), also many cascades afterwards as well.
Instead, one can make a "statistical approach":
1, take an average 1st populated state after the gamma emission of the CN*.
2, take an average of the gamma energies after the p-capture. (it includes also the photons from the CM* to the
1st state after g emission with energy of [(E_CM - Q) - E_1st_state] --> should these be excluded from the avg.
gamma energy calculation?)
3, the excited compound nucleus first decays to 1, then cascading down with the avg energy of 2,
number_of_cascades = E_1st_state/E_avg_gamma.
For the 124Xe at 7AMeV using the TALYS code the above values are:
E_1st_state = 6.74 MeV
E_avg_gamma = 2.75 MeV
number_of_cascades = 2.45
To reproduce the resulting avg. pg peak, I took a 2 and a 3 cascade simulations produced by MOCADI and mixed
them as 55% of the 2-cascade and 45% of the 3-cascade for this naiv, statistical model.
Then, to test the peakshape, I fit this mixed distribution to a part our measurement data at 7AMeV after a
sloppy Rutherford background removal (the background removal can be more improved, I used this as a quick method
for now). The calculated Chi^2/NDF ~ 3, which is not a super good value, but taking into account that this is a
strongly naiv model, and the background removal can be also improved, the chi^2/NDF value can be also
interpreted as promising. |
|
22
|
Wed Sep 25 13:03:46 2019 |
Laszlo | Analysis | simulations on 118Te+p | | I have made some simulations at 8AMeV, 7AMeV and 4AMeV energies for the main 118Te + p reaction channels. For the
Rutherford simulations credits to Yuanming! For each energy there are 3 simulations: without the scraper, with the
scraper (online), scraper (online) + E-truncation (offline). The scraper is treated as an "ideal scraper" meaning
that there is no scattering at the edges (maybe worth a GEANT4?). The slit position is at -3cm from the beam, the vertical
length is 7cm (centralized). The following channels are combined in the
simulation:
-8AMeV:
Rutherford
pg channel, 5cascade model. Photon emission is treated isotropically
pn channel, including: -->gs, -->1-->gs, -->2-->gs, -->3-->gs, -->3-->1-->gs, -->4-->1-->gs, -->5-->3-->gs,
-->5-->3-->1-->gs, -->5-->4-->1-->gs decay chains with their weights. Neutron and photon emission is treated isotropically.
-7AMeV:
Rutherford
pg channel, 4cascade model. Photon emission is treated isotropically
-4AMeV:
Rutherford
pg channel, 3cascade model. Photon emission is treated isotropically
The detector position is -2.5cm from the beam in the radial direction and centered vertically with a 45° tilt.
The cross section values for pg and pn, the pn channel mixing, and the pg cascade number is based on TALYS simulations.
The other two input parameters are the luminosity and the measurement time. For the simulation, a little
pessimistic (or realistic?) scenario is taken: L = 10^24 cm-2s-1, and t = 10^4 s. From this values, from the N=CS*L*t
equation, the pg counts (based on the TALYS
cross sections) are the following: 8AMeV ~267, 7AMeV ~163, 4AMeV ~5! This means that to reach the Gamow-window
energies is challenging, but with a well-working scraper it is not impossible to reach.
As it is visible, the 8AMeV 118Te(p,g) case is very similar to the 7AMeV 124Xe case. The p,n threshold is
somewhere ~7.5MeV. |
|
83
|
Fri Mar 20 13:21:33 2020 |
Jan, Laszlo | Analysis | 124Xe data of first night | | The unpacked data of all runs from the first night (46-50) is located at
litv-exp@lxg0188:/data.local2/E127_lmd/124Xe_1st_night.root
By taking a very quick look at the Silicon xy position histo, one can identify (please not the number of counts) clusters of hits. The x=0 and x=1 strips sees the forward scattering Rutherford particles, which remained even after scraping at 3.5cm away from the beam. This idea is supported by the simulations. Backscattered Rutherford components cannot be identified simply because of their low count rate (the exact rate should be double check). The second cluster in the middle of the detector must be our pg peak. For x>=7 strip we see the pn channel. It overlaps with the pg peak both in the position and in the energy. The energy vs Xstrip histo is NOT gainmatched, yet. Hard to make statements, but so far it seem to support the conclusions based on the position histo. The energies seem to corrsepond to the simulation: forward Rutherford, pg at same level; pn is a bit below ( ~.8% energy separation of the centers). The separation in the enregy for the backscattered Rutherford should be in the range of 2%. |
|
153
|
Sat Mar 21 23:19:14 2020 |
Jan | Analysis | 118Te52+ on H2-target - Xray spectrum 90 degree | | The attached spectrum is for the 90 degree X-ray detector taken for about 3 hours with 118Te52+.
We see two peaks at ~29 keV (K-alpha) and 44 keV (K-REC).
The resolution is about 1.5 keV, and the signal-to-background ratio is also really bad.
The increase of background events at higher E might be induced by Compton scattering of a high E gamma ray from the BaF detectors, which are active and sit close by (not anymore). |
|
159
|
Sun Mar 22 01:36:14 2020 |
Pierre-Michel | Analysis | Target overlap | | Here are the data file for the rates of the recoil detector for different values of the target bump. The average rate over background is proportional to the target overlap.
The MCP was operated at a voltage of 2500 V, where it sees mostly photons, so not really recoil ions.
Column "X" is time in seconds, Column "Content" is rate in Hz. |
|
241
|
Mon Mar 23 12:30:39 2020 |
laszlo | Analysis | with and without scraper | | |
|
244
|
Mon Mar 23 13:26:59 2020 |
Jan | Analysis | runs_90to99 SCRAPER RESULTS | | With scraper, we have roughly 150-200 (p,g) counts now.
X-rays look fine! |
|
251
|
Mon Mar 23 14:32:03 2020 |
Jan | Analysis | runs_100to104 NO-SCRAPER RESULTS | | Without scraper, we have very few (p,g) counts, hard to get numbers quickly, maybe ~50.
X-rays look fine! |
|
268
|
Wed Mar 25 15:06:09 2020 |
Laszlo | Analysis | quick summary of the run numbers | | |
|
Draft
|
Tue Apr 21 22:25:00 2020 |
Jan | Analysis | | | |
|
285
|
Fri Nov 20 15:53:37 2020 |
Jan | Analysis | Test of 2nd DSSSD (gen2) | 2021 | Here is the analysis of the test runs with the 2nd DSSD (gen2) using the quick_si_plots.c script attached.
All 32 Si channels are working with acceptable performance.
In run001, there are some additional low energy peaks in nearly all x-strips, which I do not understand yet. They are around 3 MeV and are not visible in the y-strips. It doesn't look like an electronic problem, because there are at least 4
peaks, so not a low amplitude copy of the 3 major alpha peaks between 5 - 5.8 MeV.
However, in run002, the peaks have mostly disappeared, only in x6,x7,x8,x9 is a broad structure at somewhat similar energy... maybe this has to do with the small incident angle of the alphas?
Another run to confirm and double check this would be nice. |
|
360
|
Fri May 21 01:17:37 2021 |
Laszlo | Analysis | simple analysis code | 2021 | //a simple code (template) for offline analysis
//made by Laszlo, serves as a simple demonstration for enthusiastic shifters
//it creates a "no double counting Si 2D pos" histo
//usage:
//
//save the file as eg. "simple_code.c"
//root -l
//.L simple_code.c++
//run()
//when counter finished: ".q"
#define INPUT1 "input.root"//first unpack the lmd, then give the path of the unpacked .root file.
#define OUTPUT "./"//folder of the output. minimum input: "./"
#define ROOT_NAME "dummy.root"//name of the output
#include <cmath>
#include <string>
#include <sstream>
#include <cstdlib>
#include <cstdio>
#include <ctime>
#include <fstream>
#include <iostream>
#include <stdint.h>
#include "TROOT.h"
#include "TAttText.h"
#include "TAxis.h"
#include "TCanvas.h"
#include "TChain.h"
#include "TCut.h"
#include "TF1.h"
#include "TFile.h"
#include "TGraph.h"
#include "TGraphAsymmErrors.h"
#include "TGraphErrors.h"
#include "TH1.h"
#include "TH2.h"
#include "THistPainter.h"
#include "TKey.h"
#include "TLatex.h"
#include "TLegend.h"
#include "TMath.h"
#include "TMatrixD.h"
#include "TMinuit.h"
#include "TMultiGraph.h"
#include "TNtuple.h"
#include "TPave.h"
#include "TPaveText.h"
#include "TPoint.h"
#include "TRandom.h"
#include "TRint.h"
#include "TStyle.h"
#include "TString.h"
#include "TTree.h"
#include "TH1F.h"
#include "TH2F.h"
#include "TSystem.h"
#include "TProfile.h"
#include "TVirtualFitter.h"
#include "TCanvas.h"
#include "TLegend.h"
#include "TColor.h"
#include <time.h>
using namespace std;
inline bool exists_test0 (const std::string& name) {
ifstream f(name.c_str());
return f.good();
}
///////////////////////////////////////////////////////////////////////////////////////////////
void loop(TChain *fChain){
//setting pedestal values
int PEDESTAL_LOW=400;
int PEDESTAL_HIGH=8000;
//this normally should be in a separate header, branches are defined.
UInt_t trigger;
fChain->SetBranchAddress("TRIGGER",&trigger);
UInt_t E_SiY[17];
UInt_t E_SiX[17];
UInt_t t_SiY[17];
UInt_t t_SiX[17];
for(int a=1;a<17;a++){
fChain->SetBranchAddress(Form("E_SiY%d",a),&E_SiY[a]);
fChain->SetBranchAddress(Form("E_SiX%d",a),&E_SiX[a]);
fChain->SetBranchAddress(Form("t_SiY%d",a),&t_SiY[a]);
fChain->SetBranchAddress(Form("t_SiX%d",a),&t_SiX[a]);
}
//creating histos
TH2D *h_pos_si_xy=new TH2D("h_pos_si_xy", "h_pos_si_xy",16,0.5,16.5,16,0.5,16.5);
//creating and initializing some variables used in the event loop (for "no double counting")
int r_pos_x=0,r_pos_y=0;
int dc_Ex_max=-999;
int dc_Ey_max=-999;
Long64_t nentries = fChain->GetEntries();
Long64_t nbytes = 0;
//starting the entry loop
for (Long64_t i=0; i<nentries;i++){
nbytes += fChain->GetEntry(i);
//event countdown
if ((float(i)/100000.)==int(i/100000)){cout << "event: " << i << " \tof " << nentries << endl;}
if(trigger==1){//trigger 1 = TargetON
dc_Ex_max=-999;
dc_Ey_max=-999;
for(int i_x=1;i_x<17;i_x++){
for(int i_y=1;i_y<17;i_y++){
if( ((int)t_SiX[i_x])>0 && ((int)t_SiY[i_y])>0){
if( PEDESTAL_LOW<((int)E_SiX[i_x]) && PEDESTAL_HIGH>((int)E_SiX[i_x]) &&
PEDESTAL_LOW<((int)E_SiY[i_y]) && PEDESTAL_HIGH>((int)E_SiY[i_y])){
//assign the hit to StripX and StripY where the most energy is deposited (rel. Ecalibration is needed, but roughly ok)
if(dc_Ex_max<((int)E_SiX[i_x]) && dc_Ey_max<((int)E_SiY[i_y])){
r_pos_x=i_x;
r_pos_y=i_y;
dc_Ex_max=E_SiX[i_x];
dc_Ey_max=E_SiY[i_y];
}
}
}
}
}
//Filling pos. histo
if(dc_Ex_max!=-999 && dc_Ey_max!=-999){h_pos_si_xy -> Fill(r_pos_x,r_pos_y);}
}//trigger==1
}//entry loop
//writing out the root output file
TFile *graphfile = TFile::Open((OUTPUT + (string)("") + ROOT_NAME).c_str(), "RECREATE");
graphfile -> mkdir("map");
graphfile -> cd("map");
h_pos_si_xy -> Write();
graphfile -> Close();
}//loop
//////////////////////////////////////////////////////////////////////////////////////////////////////////////
void run(){
const char *command = new char[1000];
char filename[100];
TChain *fChain = new TChain("h101");
sprintf(filename,INPUT1);
if(exists_test0(INPUT1) && exists_test0(OUTPUT)){
cout<<"\033[0;37m//loading run: "<<filename << "\033[0m" <<endl;
fChain->Add(filename);
loop(fChain);
}
else{cout<<"\033[0;31mError 404: non-existing INPUT1 or OUTPUT file!\033[0m" <<endl;}
command = "rm *.so";
gSystem->Exec(command);
command = "rm *.d";
gSystem->Exec(command);
return;
}
int main(){
run();
return(0);
} |
|
445
|
Tue May 25 11:58:30 2021 |
Jan | Analysis | 118(p,g) counts until run0115 | 2021 | The two attached plots show the collected Si data up to run0115.
Within this ~15h we have about ~70 counts in the peak region.
This translates to ~4.5 counts per hour.
And shall be compared to the 1 count/hour we had last year. |
|
469
|
Wed May 26 14:28:23 2021 |
Jan | Analysis | Xrays from 118Te & 124Xe | 2021 | Attached is a plot of the uncalibrated energy spectra taken with the 90 degree xray detector for 118Te (red) and 124Xe (blue).
Shown is the region covering the K-alpha and K-REC peaks.
Calculating the ratios (Te/Xe) for K_alpha and K-REC energies from theory one gets:
K_alpha (theory): 0.9236
K-REC (theory): 0.9308
Without energy calibration we get the following ratio (using channel numbers):
K_alpha (exp): 0.9293
K-REC (exp): 0.9344
Taking into account that the channel numbers are not calibrated, the agreement is still very good.
In summary we can be sure that we have 118Te stored at 7AMeV. |
|
523
|
Fri May 28 15:15:06 2021 |
Jan | Analysis | 118Te(p,g) counts runs 135 to 174 | 2021 | This is the summary of all Si data collected at 6 MeV/u until run174 before the Alvarez problem.
We have roughly 50-60 counts in the peak region.
This data corresponds roughly to 36hours of beam, which means ~1.5 to 2 counts/hour. |
|
572
|
Tue Jun 8 15:06:17 2021 |
Shahab | Analysis | Beam lifetime | 2021 | measured beam lifetime after the second deceleration using parallel plate Schottky detector and the RSA5103 spectrum analyser. Analysis code can be found here:
https://github.com/xaratustrah/e127_scripts/
The plots here were produced using the script:
https://nbviewer.ipython.org/github/xaratustrah/e127_scripts/blob/main/lifetime_calculator.ipynb
UPDATE 2022-06-30:
Life times have been slightly corrected. Please see attachment.
Filename Start Frame Correct Lifetime [s] Mean [s]
pgamma_lifetime-2021.05.25.21.56.37.593_on.tiq 29500 1.43
pgamma_lifetime-2021.05.25.22.02.16.618_on.tiq 29500 1.55 1.49
pgamma_lifetime-2021.05.25.22.07.22.369_off.tiq 53500 1.75
pgamma_lifetime-2021.05.25.22.13.24.787_off.tiq 34900 1.89 1.82 |
|
361
|
Fri May 21 10:00:51 2021 |
Jan | Accelerator | target bump | 2021 | The target overlap was optimized on xray and si rates.
The setting now is -10mm in SC9 |
|
413
|
Mon May 24 05:20:36 2021 |
Helmut, Yuri, Faraz, Sergey | Accelerator | FRS setup | 2021 | Logbook from start of FRS until 118Te injection |
|
438
|
Tue May 25 07:51:48 2021 |
Jan | Accelerator | ESR stacking cycle length | 2021 | The full cycle length of ESR with 30 stacks is about 7:45 minutes, out of which 15.5 sec are with target switched ON.
This is without CMB/HTD in parallel. |
|